University of Queensland researchers have used machine learning to help predict the risk of secondary bacterial infections in hospitalized COVID-19 patients. The research is published in The Lancet Microbe.
The machine learning technique can help detect whether antibiotic use is critical for patients with these infections.
Associate Professor Kirsty Short from the School of Chemistry and Molecular Biosciences said secondary bacterial infections can be extremely dangerous for those hospitalized with COVID-19.
“Estimates of the incidents of secondary bacterial infections in COVID-19 patients are broad but in some studies 100% of fatal cases have suffered a bacterial co-infection,” Dr. Short said.
“To reduce the risk of bacterial co-infections, it would be theoretically possible to just treat all COVID-19 patients with antibiotics.
“However, there’s a danger that over-treating with antibiotics could potentially lead to antibiotic resistance and the creation of bacterial superbugs.
“We’ve helped develop a robust predictive model to determine the risk of bacterial infections in COVID-19 patients, facilitating a careful use of antibiotics.”
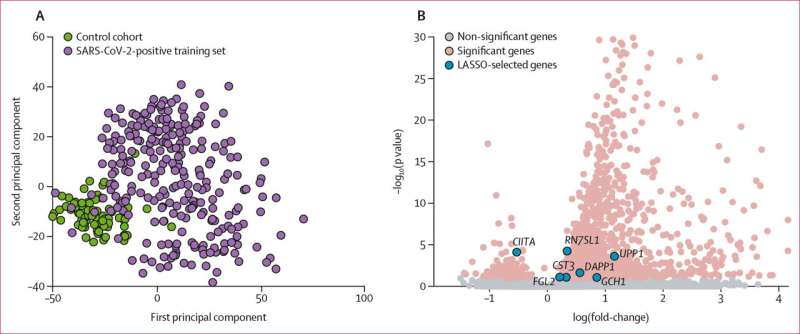
The technique is known as the “least absolute shrinkage and selection operator”—or LASSO for short.
Blood samples of COVID-19 patients from six countries were analyzed using the LASSO technique.
The team found that the expression of seven genes in a COVID-19 patient can predict their risk of developing a secondary respiratory bacterial infection after 24 hours of hospital admission.
Dr. Meagan Carney from the School of Mathematics and Physics said these seven genes will now guide clinicians to making a more informed choice when it comes to antibiotic use.
“This data raises the exciting possibility that gene transcription and analysis at the time of clinical presentation at a hospital, together with machine learning, can change the game for antibiotic prescription,” Dr. Carney said.
She also emphasized that LASSO is simplified compared to the complex machine learning methods surrounding artificial intelligence that are currently being discussed in the media.
“Research projects like this one, which use much less complex machine learning methods, can help bridge the gap between data scientists and scientists in other fields,” she said.
“We should strive towards making data science less of a black box and inspire scientists across the world to better understand how it can revolutionize the medical industry.”
The researchers would like to acknowledge the extensive international collaboration of clinicians, virologists, bioinformaticians and many other experts that made this study possible, including the PREDICT Consortium, the Snow Foundation and the Nepean Hospital.
More information:
Meagan Carney et al, Host transcriptomics and machine learning for secondary bacterial infections in patients with COVID-19: a prospective, observational cohort study, The Lancet Microbe (2024). DOI: 10.1016/S2666-5247(23)00363-4
Citation:
Using machine learning to battle COVID-19 bacterial co-infection (2024, February 2)
retrieved 2 February 2024
from https://medicalxpress.com/news/2024-02-machine-covid-bacterial-infection.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.




