Biomedical engineers at Duke University have developed a new method to improve the effectiveness of machine learning models. By pairing two machine learning models, one to gather data and one to analyze it, researchers can circumvent limitations of the technology without sacrificing accuracy.
This new technique could make it easier for researchers to use machine learning algorithms to identify and characterize molecules for use in potential new therapeutics or other materials.
The research is published in the journal Artificial Intelligence in the Life Sciences.
In traditional machine learning models, a researcher will input a dataset and the model will use that information to make predictions. While this is often effective, the abilities of these tools are limited by the datasets that are used to train them, which may often lack key information or include too much of one data type, introducing bias into the model.
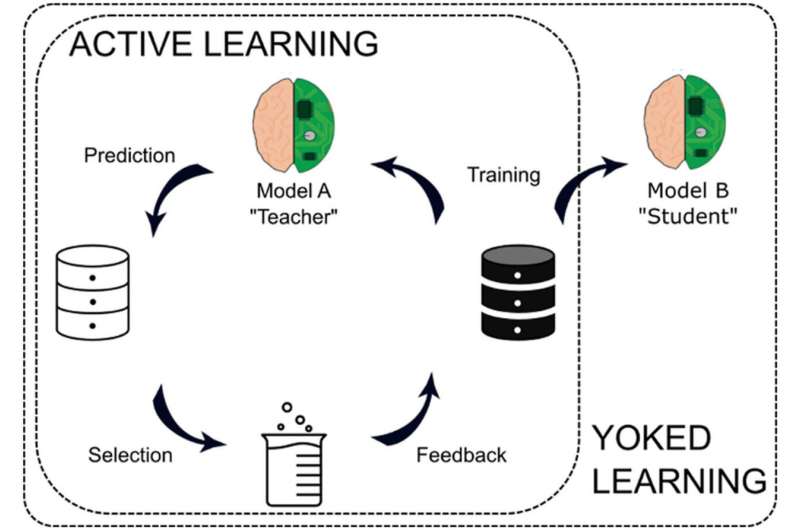
Instead, researchers have developed a technique known as active machine learning, where the model is able to ask questions or request more information if it senses a gap in the data. This questioning capability makes the model more accurate and efficient than its passive counterpart.
Although active learning is highly effective for machine learning models, the technique encounters serious limitations when it’s applied to more complex deep neural networks. Designed to mimic the human brain, these deep learning models require far more data––and computing power––than is often available, limiting their accuracy and efficacy.
Reker and his team wanted to determine if an educational concept known as “yoked learning” could be applied to the world of machine learning to improve these systems.
During yoked learning, one student will actively learn content. They can ask questions and check different textbooks for missing information. A second student will then be tasked to review the information the first student deemed as important to understand the lesson. While studies of yoked learning have shown that the second student often fails to learn concepts and retain knowledge as effectively as the actively learning student, Reker believed the technique held promise in machine learning.
“An active machine learning model knows how to go through a dataset and not only identify the important information, but also request any missing information it thinks is significant,” said Reker. “We wanted to see if we could use that active machine learning model to ‘teach’ another model using the data the actively learning model deems important.”
To understand how yoked machine learning compared to active machine learning, the team had an actively learning model identify different characteristics of molecular compounds that are important for its success as a therapeutic, such as the potential toxicity of the molecule and the molecule’s metabolism. A yoked system was developed by pairing different “teaching” machine learning models with different “student” machine learning models to identify the same characteristics based on the data selected by the “teaching” model.
The team found that while active machine learning was more accurate than the yoked system in most cases, the yoked models were very effective under certain parameters.
“We saw that the teaching model’s performance was very important for the student models,” explained Reker. “Just like in real life, an ineffective teacher means that the student isn’t set up for success. If the teaching model didn’t identify useful data, the student model wasn’t as successful at deciphering it.”
These results motivated Reker and his team to test yoked learning with a deep neural network model as the “student,” dubbed Yoked Deep Learning, or YoDeL. Unlike an active deep learning model, where the deep neural network itself is responsible for selecting the data, YoDeL has another active machine learning algorithm act as the “teacher” to actively guide the data acquisition for the deep neural network “student.”
In several comparison studies using a variety of models, the team found that their YoDeL technique either outperformed or was just as accurate as an active deep learning system when identifying different molecular characteristics. They also found that YoDeL was much faster, often only taking a few minutes to complete a task while deep active learning would take hours or even days.
The team has filed a provisional patent on the YoDeL technique, but they already have plans to continue to test and improve the parameters of the model as well as using it in the real world.
“There are lots of different machine learning and deep neural network models, so we want to determine what pairs match up really well for this yoked learning,” said Reker. “YoDeL’s ability to harness the strengths of classical machine learning models to enhance the efficacy of deep neural networks makes this a very exciting tool in a field that is always evolving. We are optimistic that we and other scientists can use this tool in the near future to help discover new drugs and new drug delivery solutions.”
More information:
Zhixiong Li et al, Yoked learning in molecular data science, Artificial Intelligence in the Life Sciences (2023). DOI: 10.1016/j.ailsci.2023.100089
Provided by
Duke University
Citation:
Machine learning models teach each other to identify molecular properties (2024, January 19)
retrieved 19 January 2024
from https://phys.org/news/2024-01-machine-molecular-properties.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.



