Data
Table 4 describes the patient populations of the datasets used in this study. Gender and race/ethnicity data and descriptors were collected from the EHR. These are generally collected either directly from the patient at registration, or by a provider, but the mode of collection for each data point was not available. Our primary dataset consisted of a corpus of 800 clinic notes from 770 patients with cancer who received radiotherapy (RT) at the Department of Radiation Oncology at Brigham and Women’s Hospital/Dana-Farber Cancer Institute in Boston, Massachusetts, from 2015 to 2022. We also created two out-of-domain test datasets. First, we collected 200 clinic notes from 170 patients with cancer treated with immunotherapy at Dana-Farber Cancer, and not present in the RT dataset. Second, we collected 200 notes from 183 patients in the MIMIC (Medical Information Mart for Intensive Care)-III database53,54,55, which includes data associated with patients admitted to the critical care units at Beth Israel Deaconess Medical Center in Boston, Massachusetts from 2001 to 2008. This study was approved by the Mass General Brigham institutional review board, and consent was waived as this was deemed exempt from human subjects research.
Only notes written by physicians, physician assistants, nurse practitioners, registered nurses, and social workers were included. To maintain a minimum threshold of information, we excluded notes with fewer than 150 tokens across all provider types. This helped ensure that the selected notes contained sufficient textual content. For notes written by all providers save social workers, we excluded notes containing any section longer than 500 tokens to avoid excessively lengthy sections that might have included less relevant or redundant information. For physician, physician assistant, and nurse practitioner notes, we used a customized medSpacy56,57 sectionizer to include only notes that contained at least one of the following sections: Assessment and Plan, Social History, and History/Subjective.
In addition, for the RT dataset, we established a date range, considering notes within a window of 30 days before the first treatment and 90 days after the last treatment. Additionally, in the fifth round of annotation, we specifically excluded notes from patients with zero social work notes. This decision ensured that we focused on individuals who had received social work intervention or had pertinent social context documented in their notes. For the immunotherapy dataset, we ensured that there was no patient overlap between RT and immunotherapy notes. We also specifically selected notes from patients with at least one social work note. To further refine the selection, we considered notes with a note date one month before or after the patient’s first social work note after it. For the MIMIC-III dataset, only notes written by physicians, social workers, and nurses were included for analysis. We focused on patients who had at least one social work note, without any specific date range criteria.
Prior to annotation, all notes were segmented into sentences using the syntok58 sentence segmenter as well as split into bullet points “•”. This method was used for all notes in the radiotherapy, immunotherapy, and MIMIC datasets for sentence-level annotation and subsequent classification.
Task definition and data labeling
We defined our label schema and classification tasks by first carrying out interviews with subject matter experts, including social workers, resource specialists, and oncologists, to determine SDoH that are clinically relevant but not readily available as structured data in the EHR, especially as dynamic features over time. After initial interviews, a set of exploratory pilot annotations was conducted on a subset of clinical notes and preliminary annotation guidelines were developed. The guidelines were then iteratively refined and finalized based on the pilot annotations and additional input from subject matter experts. The following SDoH categories and their attributes were selected for inclusion in the project: Employment status (employed, unemployed, underemployed, retired, disability, student), Housing issue (financial status, undomiciled, other), Transportation issue (distance, resource, other), Parental status (if the patient has a child under 18 years old), Relationship (married, partnered, widowed, divorced, single), and Social support (presence or absence of social support).
We defined two multilabel sentence-level classification tasks:
-
1.
Any SDoH mentions: The presence of language describing an SDoH category as defined above, regardless of the attribute.
-
2.
Adverse SDoH mentions: The presence or absence of language describing an SDoH category with an attribute that could create an additional social work or resource support need for patients:
-
Employment status: unemployed, underemployed, disability
-
Housing issue: financial status, undomiciled, other
-
Transportation issue: distance, resources, other
-
Parental status: having a child under 18 years old
-
Relationship: widowed, divorced, single
-
Social support: absence of social support
After finalizing the annotation guidelines, two annotators manually annotated the RT corpus. In total, ten thousand one hundred clinical notes were annotated line-by-line using the annotation software Multi-document Annotation Environment (MAE v2.2.13)59. A total of 300/800 (37.5%) of the notes underwent dual annotation by two data scientists across four rounds. After each round, the data scientists and an oncologist performed discussion-based adjudication. Before adjudication, dually annotated notes had a Krippendorf’s alpha agreement of 0.86 and Cohen’s Kappa of 0.86 for any SDoH mention categories. For adverse SDoH mentions, notes had a Krippendorf’s alpha agreement of 0.76 and Cohen’s Kappa of 0.76. Detailed agreement metrics are in Supplementary Tables 6 and 7. A single annotator then annotated the remaining radiotherapy notes, the immunotherapy dataset, and the MIMIC-III dataset. Table 5 describes the distribution of labels across the datasets.
The annotation/adjudication team was composed of one board-certified radiation oncologist who completed a postdoctoral fellowship in clinical natural language processing, a Master’s-level computational linguist with a Bachelor’s degree in linguistics and 1-year prior experience working specifically with clinical text, and a Master’s student in computational linguistics with a Bachelor’s degree in linguistics. The radiation oncologist and Master’s level computational linguist led the development of the annotation guidelines, and trained the Master’s student in SDoH annotation over a period of 1 month via review of the annotation guidelines and iterative review of pilot annotations. During adjudication, if there was still ambiguity, we discussed with the two Resource Specialists on the research team to provide input in adjudication.
Data augmentation
We employed synthetic data generation methods to assess the impact of data augmentation for the positive class, and also to enable an exploratory evaluation of proprietary large LMs that could not be downloaded locally and thus cannot be used with protected health information. In round 1, GPT-turbo-0301(ChatGPT) version of GPT3.5 via the OpenAI60 API was prompted to generate new sentences for each SDoH category, using sentences from the annotation guidelines as references. In round 2, in order to generate more linguistic diversity, the sample synthetic sentences output from round 1 were taken as references to generate another set of synthetic sentences. One-hundred sentences per category were generated in each round. Supplementary Table 8 shows the prompts for each sentence label type.
Synthetic test set generation
Iteration 1 for generating SDoH sentences involved prompting the 538 synthetic sentences to be manually validated to evaluate ChatGPT, which cannot be used with protected health information. Of these, after human review only 480 were found to have any SDoH mention, and 289 to have an adverse SDoH mention (Table 5). For all synthetic data generation methods, no real patient data were used in prompt development or fine-tuning.
Model development
The radiotherapy corpus was split into a 60%/20%/20% distribution for training, development, and testing respectively. The entire immunotherapy and MIMIC-III corpora were held-out for out-of-domain tests and were not used during model development.
The experimental phase of this study focused on investigating the effectiveness of different machine learning models and data settings for the classification of SDoH. We explored one multilabel BERT model as a baseline, namely bert-base-uncased61, as well as a range of Flan-T5 models62,63 including Flan-T5 base, large, XL, and XXL; where XL and XXL used a parameter efficient tuning method (low-rank adaptation (LoRA)64). Binary cross-entropy loss with logits was used for BERT, and cross-entropy loss for the Flan-T5 models. Given the large class imbalance, non-SDoH sentences were undersampled during training. We assessed the impact of adding synthetic data on model performance. Details on model hyper-parameters are in Supplementary Methods.
For sequence-to-sequence models, input consisted of the input sentence with “summarize” appended in front, and the target label (when used during training) was the text span of the label from the target vocabulary. Because the output did not always exactly correspond to the target vocabulary, we post-processed the model output, which was a simple split function on “,” and dictionary mapping from observed miss-generation e.g., “RELAT → RELATIONSHIP”. Examples of this label resolution are in Supplementary Methods.
Ablation studies
Ablation studies were carried out to understand the impact of manually labeled training data quantity on performance when synthetic SDoH data is included in the training dataset. First, models were trained using 10%, 25%, 40%, 50%, 70%, 75%, and 90% of manually labeled sentences; both SDoH and non-SDoH sentences were reduced at the same rate. The evaluation was on the RT test set.
Evaluation
During training and fine-tuning, we evaluated all models using the RT development set and assessed their final performance using bootstrap sampling of the held-out RT test set. Bootstrap sample number and size were calculated to achieve a precision level for the standard error of macro F1 of ±0.01. The mean and 95% confidence intervals from the bootstrap samples were calculated from the resulting bootstrap samples. We also sampled to ensure that our standard error on the 95% confidence interval limits was <0.01 as follows: Our selected bootstrap sample size matched the test data size, sampling with replacement. We then computed the 5th and 95th percentile values for each of the calculated k samples from the resulting distributions. The standard deviation of these percentile values was subsequently determined to establish the precision of the confidence interval limits. Examples of the bootstrap sampling calculations are in Supplementary Methods.
For each classification task, we calculated precision/positive predictive value, recall/sensitivity, and F1 (harmonic mean of recall and precision) as follows:
-
Precision = TP/(TP + FP)
-
Recall = TP/(TP + FN)
-
F1 = (2*Precision*Recall)/(Precision+Recall)
-
TP = true positives, FP = false positives, FN = false negatives
Manual error analysis was conducted on the radiotherapy dataset using the best-performing model.
ChatGPT-family model evaluation
To evaluate ChatGPT, the Scikit-LLM65 multilabel zero-shot classifier and few-shot binary classifier were adapted to form a multilabel zero- and few-shot classifier (Fig. 4). A subset of 480 synthetic sentences whose labels were manually validated, were used for testing. Test sentences were inserted into the following prompt template, which instructs ChatGPT to act as a multilabel classifier model, and to label the sentences accordingly:
Sample target: [LABELS]”
[TEXT] was the exemplar from the development/exemplar set.
[LABELS] was a comma-separated list of the labels for that exemplar, e.g. PARENT,RELATIONSHIP.
Example of prompt templates used in the SKLLM package for GPT-turbo-0301 (GPT3.5) and GPT4 with temperature 0 to classify our labeled synthetic data. {labels} and {training_data} were sampled from a separate synthetic dataset, which was not human-annotated. The final label output is highlighted in green.
Of note, because we were unable to generate high-quality synthetic non-SDoH sentences, these classifiers did not include a negative class. We evaluated the most current ChatGPT model freely available at the time of this work, GPT-turbo-0613, as well as GPT4–0613, via the OpenAI API with temperature 0 for reproducibility.
Language model bias evaluation
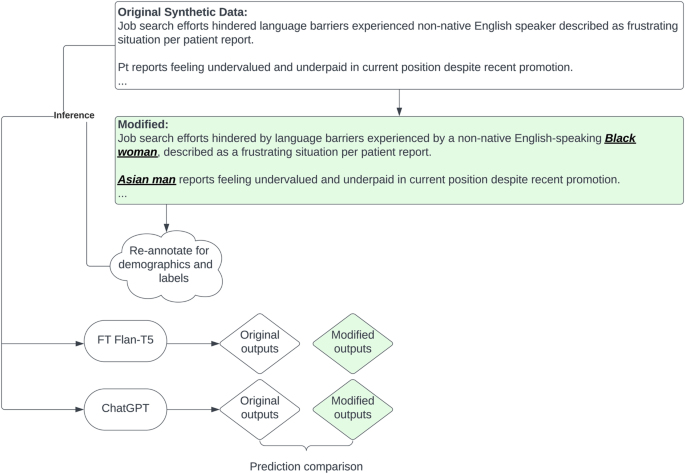
In order to test for bias in our best-performing models and in large LMs pre-trained on general text, we used GPT4 to insert demographic descriptors into our synthetic data, as illustrated in Fig. 5. GPT4 was supplied with our synthetically generated test sentences, and prompted to insert demographic information into them. For example, a sentence starting with “Widower admits fears surrounding potential judgment…” might become “Hispanic widower admits fears surrounding potential judgment…”. The prompt was as follows (in a batch of 10 ensure demographic variations):
“role”: “user”, “content”: “[ORIGINAL SENTENCE]\n swap the sentences patients above to one of the race/ethnicity [Asian, Black, white, Hispanic] and gender, and put the modified race and gender in bracket at the beginning like this \n Owner operator food truck selling gourmet grilled cheese sandwiches around town => \n [Asian female] Asian woman owner operator of a food truck selling gourmet grilled cheese sandwiches around town”
[ORIGINAL SENTENCE] was a sentence from a selected subset of our GPT3.5-generated synthetic data
These sentences were then manually validated; 419 had any SDoH mention, and 253 had an adverse SDoH mention.
Comparison with structured EHR data
To assess the completeness of SDoH documentation in structured versus unstructured EHR data, we collected Z-codes for all patients in our test set. Z-codes are SDoH-related ICD-10-CM diagnostic codes, mapped most closely with our SDoH categories present as structured data for the radiotherapy dataset (Supplementary Table 9). Text-extracted patient-level SDoH information was defined as the presence of one or more labels in any note. We compared these patient-level labels to structured Z-codes entered in the EHR during the same time frame.
Statistical analysis
Macro-F1 performance for each model type was compared when developed with or without synthetic data and for the ChatGPT-family model comparisons using the Mann–Whitney U test. The rate of discrepant SDoH classifications with and without the injection of demographic information was compared between the best-performing fine-tuned models and ChatGPT using chi-squared tests for multi-class comparisons and 2-proportion z tests for binary comparisons. A two-sided P ≤ 0.05 was considered statistically significant. Statistical analyses were carried out using the statistical Python package in scipy (Scipy.org). Python version 3.9.16 (Python Software Foundation) was used to carry out this work.