Patient selection
In the discovery set for high-throughput proteomics analysis, 29 patients with BC diagnosed between 1998 and 2014 at Seoul National University Hospital (SNUH) with available clinical follow-up data including DFS, MFS, and OS were included. We defined the LM as the development of metastasis after 5 years from the diagnosis.
In addition, a retrospective consecutive cohort of 1002 patients with BCs who underwent surgical resection and were diagnosed between 2009 and 2012 in SNUH were recruited. Patients who received neoadjuvant chemotherapy were excluded. FFPE surgical samples archived in the Department of Pathology, SNUH, TMA were constructed. The experienced breast pathologist (H.S.R) selected the 2 mm-sized representative areas from H&E slides of the samples, and the TMAs were made (Superbiochips Laboratory, Seoul, Republic of Korea).
This study was approved by the Institutional Review Board (IRB) of SNUH, and the individual informed consent forms from the patients were waived by the decision of IRB. We confirm that all methods were performed in accordance with the relevant guidelines and regulations.
High-throughput proteomics assay
From the FFPE blocks of surgically resected samples of the discovery set, 10 μm thick sections were deparaffinized in xylene twice. The protein uses a combination of acetone precipitation and filter-aided sample preparation (FASP)32. Desalted pooled peptides were fractionated using the stage tip-based high-pH peptide fractionation method32.
The pre-fractionated peptides were analyzed on an LC–MS system with an Easy-nLC 1000 (Thermo Fisher Scientific, Waltham, MA, USA) equipped with a nanoelectrospray ion source (Thermo Fisher Scientific) and Q-Exactive mass spectrometer (Thermo Fisher Scientific). The peptide samples were separated into a trap column and an analytical column (Thermo Fisher Scientific).
Raw MS/MS files were processed with MaxQuant (version 1.6.1.0) using the Andromeda search engine against the Human Uniprot protein sequence database (December_2014, 88 657 entries). Experiment details are presented in the Supplementary Methods.
Proteomics data analysis
Preprocessing
Protein levels were normalized and statistical analyses were performed by Perseus (version 1.5.8.5). ANOVA was used to determine DEP among NM, LM, and M groups. Peptide intensity data from tandem mass spectrometry (MS/MS) were processed using MaxQuant acquired. LFQ intensity was used as a protein quantification measure. The proteome dataset of breast cancer metastasis was preprocessed as follows. Only proteins that were identified in all replicates were used. Missing values were predicted by k-nearest neighbor imputation. As a result, in total 9455 proteins were quantified and resulted in 6639 proteins after filtration and imputation. The MS-based proteomics data of all identified peptides has been registered in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (data set identifier: PXD015171).
Machine learning-based feature selection
The goal is to select proteins as features for biomarker candidates whose abundance level can discriminate metastasis types: NM, M, and LM. The sample size of each metastasis type is 9 for NM, 10 for M, and 10 for LM, respectively.
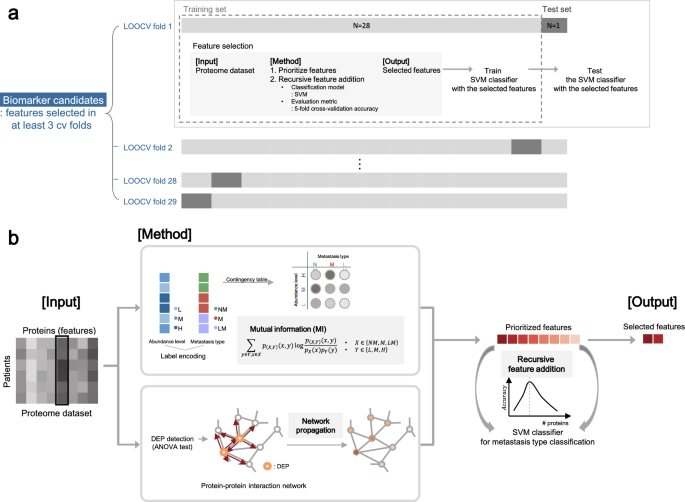
The full schematic overview of the biomarker candidate selection method is illustrated in Fig. 5 and the pseudo-code of the algorithm is in Supplementary Fig. S6. Our feature selection method operates in two hierarchical levels. The higher level is to choose protein features that are selected multiple times in the 29 LOOCV experiments. On the lower level of each of 29 LOOCV experiments, a set of features is selected in the recursive feature addition scheme. Below we explain how the algorithm works.
Schematic overview of biomarker candidate selection. (A) Leave-one-out cross-validation (LOOCV) scheme for biomarker candidate selection. For each CV fold, a sample was used as a held-out test set, while the other samples were used for feature selection. After feature selection, SVM classifier was trained with the selected features and tested with the test set. (B) Within each LOOCV training set, we prioritized proteins by mutual information (MI) network propagation (NP). Proteins were ranked in terms of the discriminative power of metastasis type measured by MI. After feature prioritization, features were added to the classification model via recursive feature selection, until the accuracy reached the maximum.
At a higher level (Fig. 5A, Supplementary Fig. S6; line 2–5, 23–25), we used the LOOCV scheme for biomarker candidate selection to identify protein sets whose abundance can discriminate metastasis type of unseen patients. For each CV fold, a sample was used as a held-out test set (Supplementary Fig. S6; line 4), while the other samples were used as a training set for feature selection (Supplementary Fig. S6; line 5), denoted as tr_LOOCV. In each of 29 LOOCV folds, we performed feature selection by recursive feature addition and metastasis prediction tasks in fivefold CV settings. To evaluate the selected features, SVM classifier was trained with the selected features and tested with the test set in terms of precision, recall, and F1 score. We reported macro averaged evaluation metrics of precision, recall and F1 score respectively, which is to calculate an evaluation metric for all classes individually and average them. As a result, a different feature set is selected and tested for each of 29 LOOCV folds. Thus, the final features are features that are selected multiple times, 3 times, out of 29 LOOCV folds. Since we are selecting features out of 6639 protein features that are measured by mass spectrometry experiments, this is a very stringent feature selection criteria. Statistically, we can measure p-values for features selected as features at least 3 times as \(1.95\times {10}^{-6}\).
At the lower level (Fig. 5B), in each of the 29 LOOCV folds, protein features were selected by performing recursive feature addition (Supplementary Fig. S6; line 6–22). The first step is to prioritize proteins for the recursive feature addition analysis using all samples in tr_LOOCV (Supplementary Fig. S6; line 6). Proteins were ranked in terms of the discriminative power of metastasis type measured by MI. When there were ties for multiple proteins, we used the NP method to break the ties. The second step is to select features by recursive feature addition (Supplementary Fig. S6; line 7–22). Specifically, protein features were recursively added to an SVM classifier according to the rank until the prediction performance of the classifier reached peak. The prediction performance is measured with fivefold cross-validation within a tr_LOOCV (Supplementary Fig. S6; line 10–17). The feature set where prediction performance is maximum was regarded as the final selected feature for each LOOCV fold (Supplementary Fig. S6; line 18–21).
Feature prioritization step
MI is a metric for statistical dependency between two variables, the metastasis label and protein abundance group in this case. Relevance between protein abundance level and metastasis type is computed as below.
$$\sum_{y\in Y}\sum_{x\in X}{p}_{\left(X,Y\right)}\left(x,y\right){\text{log}}\frac{{p}_{\left(X,Y\right)}\left(x,y\right)}{{p}_{X}\left(x\right){p}_{Y}\left(y\right)}.$$
Discrete variable \(X\in \{NM,M,LM\}\) denotes metastasis type and the other discrete variable \(Y\in \{L,M,H\}\) denotes protein abundance level, while \(L\) encodes for low abundance level, \(M\) for medium abundance level, and \(H\) for high abundance level. Since raw LFQ protein abundance is a continuous variable, we transformed the variable into a multi-chotomous discrete variable \(X\) where protein abundance levels are categorized into 3 groups, where \(L\) for low, \(M\) for medium, and \(H\) for high abundance level, respectively. To handle the label imbalance issue, we sampled 100 times with an equivalent sample size (n = 9) for each metastasis type. Averaged MI across sampling batches was used to prioritize proteins.
To prioritize features in terms of influence in a biological network, NP simulates the effects of biomarkers in a biological network. In detail, NP re-prioritized proteins according to the relevance with the seed proteins, by propagating the influence of seed proteins along the PPI given a STRING (v11.0)33 as a protein–protein interaction network. For seed selection, DEPs among the three metastasis types were detected with the ANOVA test, for proteins with adjusted p-value < 0.1 after Benjamini–Hochberg multiple test correction. For a network, STRING network was trimmed with a confidence score, where normalized edge confidence < 0.5 was filtered out. For the implementation of the network propagation algorithm, we used HotNet234.
Feature selection step
After feature prioritization, features were recursively added to the classification model according to the feature rank. For evaluating each iteration of the recursive feature addition, SVM with an RBF kernel was trained to discriminate the metastasis types by recursively adding features according to the rank. The SVM classifier with RBF kernel was chosen as a classifier because the model showed the highest prediction performance compared to the other classifiers (Supplementary Table S1). As a new feature was added, the prediction performance, i.e., accuracy, of the current classification model in each iteration was computed in a threefold cross-validation scheme with a training set. Accuracy is calculated as macro averaged accuracy for multiclass classification35. Feature addition was iteratively performed until the accuracy reached the maximum.
Cross validation using publicly available gene expression dataset
To ascertain the validity of the candidate biomarker derived from the proteomics assay, we used the publicly available gene expression dataset—METABRIC36—to assess the prognostic significance of biomarker expression. Kaplan–Meier survival analyses according to VWA5A gene expression level was done via web-based analytic platform, CTGS (http://ctgs.biohackers.net/)37 accessed at June 2020.
Cell culture and invasion/migration assay
We used HR+ cell lines (T47D, MCF7), HER2+ cell lines (HCC1954, JIMT-1, and SKBR3), TNBC cell lines (BT20, HCC70, HCC1143, HCC1937, MDA-MB-468, MDA-MB-231 and HCC 1395) for in vitro experiments. All cell lines were obtained from the Korean Cell Line Bank (KCLB, Seoul, Republic of Korea). T47D and HCC70 were cultured in RPMI (Gibco, Carlsbad, CA, USA), and BT20 was cultured in DMEM (Gibco) containing 10% fetal bovine serum (FBS; Gibco) and 1% penicillin and streptomycin (Gibco). The cells were maintained at 37 °C in a humidified atmosphere of 95% air and 5% CO2 and were screened periodically for mycoplasma contamination.
siRNAs targeting VWA5A were generated and purchased from Bioneer (Daejeon, Republic of Korea). T47D, BT20 and HCC700 cells were transfected with siVWA5A using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. After an incubation period of 48 h, mRNA expressions of each biomarker were assessed by RT-PCR and compared with the controls. RNA from the cells was isolated using TRIzol (Invitrogen), and synthesized respective cDNA was amplified using specific primers and HIPI plus Master mix (ElpisBio, Daejeon, Republic of Korea).
Cell migration and invasion assays were performed using 24-well inserts (Corning Incorporated, NY, USA) with 8-μm pores according to the manufacturer’s instructions. For transwell migration assays, 5 × 105 cells were inoculated into the upper chamber, while culture medium containing 10% FBS was added into the lower chamber. After 24 h of incubation, the cells on the top of the membrane were removed, and the migrant cells were washed with phosphate-buffered saline (PBS), stained by 1% crystal violet for 10 min, and counted in 3 randomly selected fields under the microscope (Nikon, Tokyo, Japan). The experiments were replicated three times each.
For Matrigel invasion assay, the upper wells of Boyden chambers (Corning) were coated with 2 mg/ml of Matrigel at 37 °C incubator with 5% CO2. 5 × 105 cells were put into the upper chamber, with 10% FBS added medium in the lower chamber. Invaded cells, after 24 h of incubation, were processed and counted in triplicate, as described above.
Analysis of cell proliferation
To measure cell proliferation rate depending on VWA5A gene expression, cells were seeded at 1 × 104 cells per well in a 96-well plate. Cell proliferation was determined using the Cell Counting Kit-8 (Dojindo Laboratoried, Japan) every day for 3 days.
Immunoblotting
Whole proteins of breast cell lines were extracted using T-PER (Pierce, Rockford, IL) containing a cocktail of protease inhibitors (Roche). Proteins were detected using standard immunoblotting procedures and the appropriate primary antibodies. Anti-BCSC was purchased from Novus Biologicals (Centinnial, CO) and anti-β-actin was purchased from Santa Cruz Biotechnology (Dallas, TX).
qPCR
Cultured cells were lyzed in Trizol (Takara, Japan) and total intracellular RNAs were extracted according to the manufacturer’s instructions. cDNA was generated using M-MLV reverse transcriptase (Promega, Madison, WI) and random primers (Promega). Relative quantitative real-time PCR was performed using GO Taq® qPCR Master Mix (Promega) and analyzed on a CFX96™ Real-Time System (Bio-Rad, Hercules, CA).
Immunohistochemistry
From the TMA block of the validation set, 4 μm thick sections were taken to perform IHC for VWA5A. Monoclonal anti-VWA5A antibody (dilution 1:400; clone OTI3D6; Novus Biologicals, Centennial, CO, USA) was used, and immunostaining was performed using Benchmark automatic immunostaining device (Ventana BenchMark XT Staining System, Tucson, AZ) following the manufacturer’s guidelines. Interpretation of the IHC slides were done by a breast pathologist (K.J.) using histoscore (H-score) while blinded to the clinicopathological characteristics. H-score of 50 was used as a cut-off value discriminating VWA5A-high and VWA5A-low.
Statistical analysis
We used the chi-square test and linear-by-linear tests to compare categorical variables and the Kruskal–Wallis test to compare continuous variables, as appropriate. Kaplan–Meier survival analyses were performed based on the log-rank method. Harrell’s c-index was used to assess the discriminating ability of biomarker expression. Statistical significance was defined as the p-value less than 0.05. All analyses were performed by SPSS software (version 25; IBM SPSS Statistics, IBM Corporation, Armonk, NY, USA) and R version 3.6.3 (www.r-project.org).
Ethics declarations and informed consent statement
This study was approved by the Institutional Review Board (IRB) of SNUH (IRB No. 1612-011-811), and the individual consent forms from the patients were waived by the decision of IRB.