ChEMBL provides multitask prediction model from its github repo. And shared useful blog post.
https://chembl.blogspot.com/2019/05/multi-task-neural-network-on-chembl.html
By using the code, we can get predicted target list from given molecules. And the prediction can run python, C++, JS and Knime! ChEMBL team provides not only source code but also predicted results when we search compound in ChEMBL DB. The screen shot is shown below.
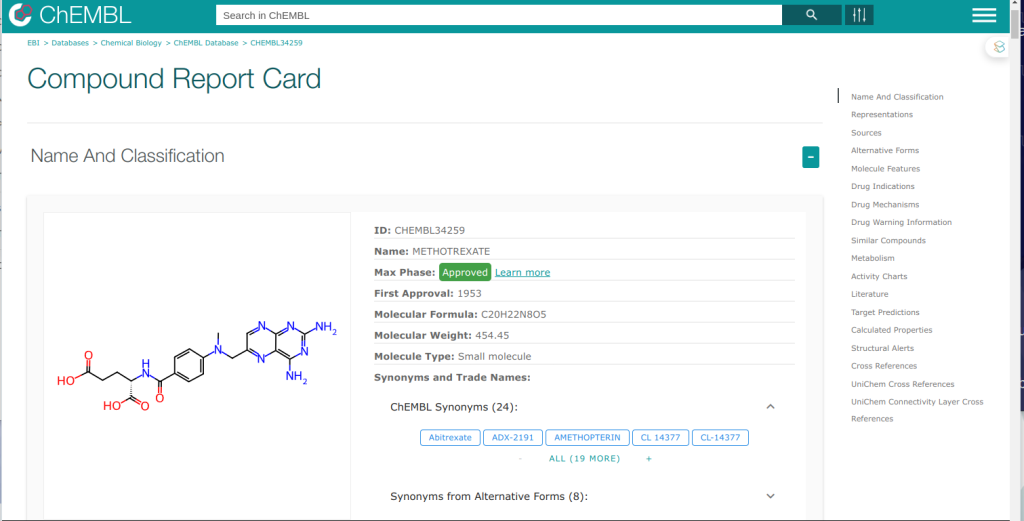

I found the bottom of the predicted target list, prediction API is provided too! For example, I can get predicted results as JSON format when I posted query below.
curl 'https://www.ebi.ac.uk/chembl/target-predictions' -H 'Content-Type: application/json' --data-raw '{"smiles":"CN(Cc1cnc2nc(N)nc(N)c2n1)c1ccc(C(=O)N[C@@H](CCC(=O)O)C(=O)O)cc1"}'
Web based API is useful because user don’t need prepare the environment on own computational enviroment. So I tried to make query with python requests, because by using python I can make chemoinformatics pipeline easily.
Here is an example code 😉
Tips for making query is data definition part.
F-String can’t escape curry branckets with backslash. {{someting}} means {someting}, as a normal strings.
### tips
data = f'{{"smiles":"{smi}"}}' >>> '{"smiles" : "user defined smiles"}'
The code worked well but I wondered that predicted results don’t contain COX2 which is primary target of celecoxib. I would like to check some other approved drugs with same way.





