General sequence-to-sequence NERRE
We fine-tune Llama-2 and GPT-3 models to perform NERRE tasks using 400−650 manually annotated text-extraction (prompt-completion) pairs. Extractions contain the desired information formatted with a predefined, consistent schema across all training examples. These schemas can range in complexity from English sentences with predefined sentence structures to lists of JSON objects or nested JSON objects. In principle, many other potential schemas (e.g., YAML, psuedocode) may also be valid, though we do not explore those here. Once fine-tuned on sufficient data adhering to the schema, a model will be capable of performing the same information extraction task on new text data with high accuracy. The model outputs completions in the same schema as the training examples. We refer to this approach generally as “LLM-NERRE”.
Our general workflow for training GPT-3 and Llama-2 to perform NERRE tasks is outlined in Fig. 2. Annotations are performed by human domain experts to create an initial training set, and then a partially trained model (GPT-3) is used to accelerate the collection of additional training examples. Fine-tuning is then performed on these examples to produce a “partially trained” model, which is used to pre-fill annotations that are subsequently corrected by the human annotator before being added to the training set. Once a sufficient number of annotations have been completed, the final fine-tuned model is capable of extracting information in the desired format without human correction. Optionally, as illustrated in Figs. 5 and 6, the structured outputs may be further decoded and post-processed into hierarchical knowledge graphs.
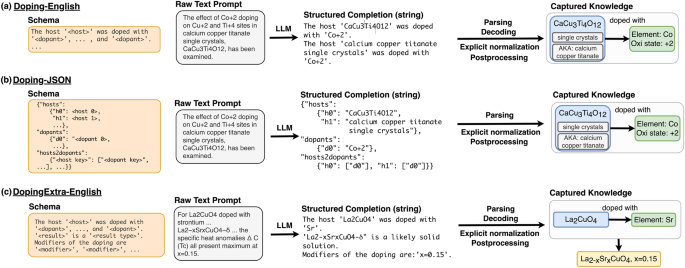
In all three panels, an LLM trained to output a particular schema (far left) reads a raw text prompt and outputs a structured completion in that schema. The structured completion can then be parsed, decoded, and formatted to construct relational diagrams (far right). We show an example for each schema (desired output structure). Parsing refers to the reading of the structured output, while decoding refers to the programmatic (rule-based) conversion of that output into JSON form. Normalization and postprocessing are programmatic steps which transform raw strings (e.g., “Co+2″) into structured entities with attributes (e.g., Element: Co, Oxidation state +2). a Raw sentences are passed to the model with Doping-English schema, which outputs newline-separated structured sentences that contain one host and one or more dopant entities. b Raw sentences are passed to a model with Doping-JSON schema, which outputs a nested JSON object. Each host entity has its own key-value pair, as does each dopant entity. There is also a list of host2dopant relations that links the corresponding dopant keys to each host key. c Example for the extraction with a model using the DopingExtra-English schema. This first part of the schema is the same as in a, but additional information is contained in doping modifiers, and results-bearing sentences are included at the end of the schema.
Task and schema design
Solid-state impurity doping schema
The Doping-English and Doping-JSON schemas aim to extract two entity types (host and dopant) and the relations between them (host–dopant), returned as either English sentences or a list of one or more JSON objects. Hosts are defined as the host crystal, sample, or material class along with crucial descriptors in its immediate context (e.g., “ZnO2 nanoparticles”, “LiNbO3″, “half-Heuslers”). Dopants are taken to be any elements or ions that are minority species, intentionally added impurities, or specific point defects or charge carriers (“hole-doped”, “S vacancies”). One host may be doped with more than one dopant (e.g., separate single-doping or co-doping), or the same dopant may be linked to more than one host material. There may also be many independent pairs of dopant-host relations, often within a single sentence, or many unrelated dopants and hosts (no relations). We impose no restriction on the number or structure of the dopant-host relations beyond that each relation connects a host to a dopant. The Doping-JSON schema represents the graph of relationships between hosts and dopants within a single sentence, where unique keys identify dopant and host strings. The model aims to learn this relatively loose schema during fine-tuning. A separate key, “hosts2dopants”, describes the pairwise relations according to those unique keys. The Doping-English schema encodes the entity relationships as quasi-natural language summaries. The Doping-English schema represents the same information as the Doping-JSON schema, but more closely mimics the natural language pre-training distribution of the LLMs we tested. When there are multiple items to extract from the same sentence, the output sentences are separated by newlines.
For the DopingExtra-English schema, we introduce two additional entities: modifiers and result, without explicit linking (i.e., NER only). The results entity represents formulae with algebra in the stoichiometric coefficients such as AlxGa1−xAs, which are used for experiments with samples from a range of compositions or crystalline solid solutions (e.g., CaCu3−xCoxTi4O12). We also include stoichiometries where the algebra is substituted (i.e., x value specified) and the doped result is a specific composition (e.g., CaCu2.99Co0.1Ti4O12). Modifiers are loosely bounded entity encapsulating other descriptors of the dopant-host relationship not captured by dopant, host, or result. These can be things like polarities (e.g., “n-type”, “n-SnSe”), dopant quantities (e.g., “5 at.%”, “x < 0.3″), defect types (e.g., “substitutional”, “antisite”, “vacancy”) and other modifiers of the host to dopant relationship (e.g., “high-doping”, “degenerately doped”). These entities (host, dopant, result, and modifiers) were chosen to define a minimal effective schema for extracting basic doping information.
All doping-related models are trained to work only on single sentences. The main motivation for this design choice is that the vast majority of dopant-related data can be found within single sentences, and the remaining relational data is often difficult to resolve consistently for both human annotators and models. We expand on problems with annotations and ambiguity in Supplementary Discussion 5 and we further explain the doping task schemas in Supplementary Note 1.
General materials information schema
In our previous work4,5, we focused on NER for a specific set of entity types that are particularly relevant in materials science: materials, applications, structure/phase labels, synthesis methods, etc. However, we did not link these labeled entities together to record their relations beyond a simple “bag-of-entities” approach. In this work, we train an LLM to perform a “general materials information extraction” task that captures both entities and the complex network of interactions between them.
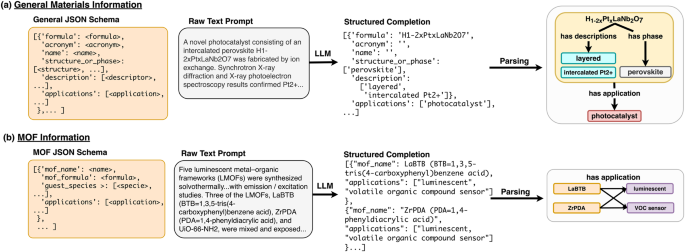
The schema we have designed for this task encapsulates an important subset of information about solid compounds and their applications. Each entry in the list, a self-contained JSON document, corresponds one-to-one with a material mentioned in the text. Materials entries are ordered by appearance in the text. The root of each entry starts with a compound’s name and/or its chemical formula. If a name or formula is not mentioned for a material, no information about that material is extracted from the text. We also extract acronyms mentioned for a material’s name/formula, although in cases where only an acronym is mentioned we do not create a material entry for the compound. Compounds that are not solids (ions, liquids, solvents, solutions, etc) are generally not extracted. The name, formula, and acronym fields are exclusively given string value in the JSON document for each material whereas the description, structure_or_phase, and applications fields are lists of an arbitrary number of strings. We label this model General-JSON, and an example is shown in Fig. 6 (a).
In both panels, an LLM trained using a particular schema (desired output structure, far left) is prompted with raw text and produces a structured completion as JSON. This completion can then be parsed to construct relational diagrams (far right). Each task uses a different schema representing the desired output text structure from the LLM. a Schema and labeling example for the general materials-chemistry extraction task. Materials science research paper abstracts are passed to an LLM using General-JSON schema, which outputs a list of JSON objects representing individual material entries ordered by appearance in the text. Each material may have a name, formula, acronym, descriptors, applications, and/or crystal structure/phase information. b Schema and labeling example for the metal-organic frameworks extraction task. Similar to the General-JSON model, the MOF-JSON model takes in full abstracts from materials science research papers and outputs a list of JSON objects. In the example, only MOF name and application were present in the passage, and both MOFs (LaBTB and ZrPDA) are linked to both applications (luminescent and VOC sensor).
Description entities are defined as details about a compound’s processing history, defects, modifications, or the sample’s morphology. For example, consider the hypothetical text “Pt supported on CeO2 nanoparticles infused with Nb…”. In this case, the description value for the material object referring to “Pt” might be annotated as “[‘supported on CeO2’]”, and the description entities listed for “CeO2″ would be “[‘nanoparticles’, ‘Nb-doped’]”.
Structure_or_phase entities are defined as information that directly implies the crystalline structure or symmetry of the compound. Crystal systems such as “cubic” or “tetragonal”, structure names such as “rutile” or “NASICON”, and space groups such as “Fd3m” or “space group No. 93″ are all extracted in this field. We also include any information about crystal unit cells, such as lattice parameters and the angles between lattice vectors. “Amorphous” is also a valid structure/phase label.
Applications are defined as high-level use cases or major property classes for the material. For example, a battery cathode material may have “[‘Li-ion battery’, ‘cathode’]” as its applications entry. Generally, applications are mentioned in the order they are presented in the text, except for certain cases such as battery materials, in which case the type of device is generally mentioned before the electrode type, and catalysts, where the reaction catalyzed is generally listed following the “catalyst” entity in the list (e.g.,“[‘catalyst’, ‘hydrogenation of citral’]”).
More details about the general materials information task schema are provided in the Supplementary Discussion 4.
Metal–organic framework (MOF) schema
The schema used for the MOF cataloging task is based on the general materials information schema described in the previous section, which was modified to better suit the needs of MOF researchers. We developed this schema to extract MOF names (name), an entity for which there is no widely accepted standard60, and chemical formulae (formula), which form the root of the document. If no name or formula is present, no information is extracted for that instance. In addition, because there is a great deal of interest in using MOFs for ion and gas separation61,62, we extract guest species, which are chemical species that have been incorporated, stored, or adsorbed in the MOF. We extract applications the MOF is being studied for as a list of strings (e.g., “[‘gas-separation’]” or “[‘heterogeneous catalyst’, ‘Diels-Alder reactions’]”) as well as a relevant description for the MOF, such as its morphology or processing history, similar to the general information extraction schema. Entries in the list are generally added in the order the material names/formulae appear in the text. The MOF extraction model is labeled MOF-JSON, and an example is shown in Fig. 6 (b).
Comparison baselines and evaluation
To compare our model with other sequence-to-sequence approaches to information extraction, we perform a benchmark of two methods on the doping task to compare to the LLM-NERRE models. The first employs the seq2rel method of Giorgi et al.41 for the host-dopant task. We formatted host-dopant relationships under tags labeled @DOPANT@ and @BASEMAT@ (base/host material), with their relationship signified by @DBR@ (“dopant-base material relationship”); these sequences were constructed from the same training data as the Doping-JSON and Doping-English models. We trained seq2rel to perform sentence-level extraction with 30 epochs, batch size of 4, encoder learning rate 2 × 10−5, decoder learning rate 5 × 10−4, and pretrained BiomedNLP BERT tokenizer51 (further training details can be found in the Supplementary Note 4). Additionally, we compare against the previously published MatBERT doping-NER model5 combined with proximity-based heuristics for linking (see Supplementary Note 5). With this method, a MatBERT NER model pretrained on ~50 million materials science paragraphs and fine-tuned on 455 separate manually annotated abstracts first extracts hosts and dopants and then links them if they co-occur in the same sentence.
Datasets
Datasets were prepared from our database of more than 8 million research paper abstracts63. Annotations were performed by human annotators using a graphical user interface built using Jupyter64, although in principle annotations could be conducted via a simple text editor. To accelerate the collection of training data, new annotations are collected via a “human in the loop” approach where models are trained on small datasets and their outputs are used as starting points and corrected by human annotators (see Fig. 2.) This process of training and annotation is completed multiple times until a sufficiently large set of training data was achieved. Each dataset was annotated by a single domain expert annotator. Class support for each annotated dataset is provided in Supplementary Tables 1–3.
Doping dataset
Training and evaluation data was gathered from our database of research paper abstracts using the keywords “n-type”, “p-type”, “-dop”, “-codop”, “doped”, “doping”, and “dopant” (with exclusions for common irrelevant keywords such as “-dopamine”), resulting in ~375k total abstracts. All doping tasks were trained on text from 162 randomly selected abstracts, comprising 1215 total sentences and filtered with regular expressions to only include 413 relevant (potentially including doping information) sentences. Doping tasks were tested on an additional 232 sentences (77 relevant by regex) from a separate holdout test set of 31 abstracts.
General materials dataset
Training and evaluation data was gathered from our abstract database by using keywords for a variety of materials properties and applications (e.g., “magnetic”, “laser”, “space group”, “ceramic”, “fuel cell”, “electrolytic”, etc). For each keyword a materials science domain expert annotated ~10–50 abstracts, which resulted in ≈650 entries manually annotated according to the general materials information schema. Results were evaluated using a 10% random sample for validation, and this procedure was averaged over five trials using different random train/validation splits with no hyperparameter tuning.
Metal–organic framework dataset
Training and evaluation data was selected from our database using the keywords “MOF”, “MOFs”, “metal-organic framework”, “metal organic framework”, “ZIF”, “ZIFs”, “porous coordination polymer”, and “framework material”, which produced approximately 6,000 results likely containing MOF-related information. From these, 507 abstracts were randomly selected and annotated by a MOF domain expert. Results were evaluated using the same repeated random split procedure as the general materials dataset in the previous section.
GPT-3 fine tuning details
For all tasks, we fine-tune GPT-3 (‘davinci’, 175B parameters)26 using the OpenAI API, which optimizes the cross-entropy loss on predicted tokens. Doping models were trained for 7 epochs at a batch size of 1, with inference temperature of 0 and output limited to a maximum length of 512 tokens (all doping models) or 1024 tokens (General-JSON, MOF-JSON). The intermediate models shown in Fig. 4 were trained with a number of epochs depending on the number of training samples t: 2 epochs for 20≤t < 26, 4 epochs for 26 < t ≤ 27, and 7 epochs for t ≥ 28. Models for the MOF and general materials extraction tasks were trained for 4 epochs with a batch size of 1. We use a learning rate multiplier of 0.1 and a prompt loss weight of 0.01 but have not performed hyperparameter tuning for these hyperparameters. For all tasks, the start and end tokens used were “\n\n\n###\n\n\n” and “\n\n\nEND\n\n\n”.
Llama-2 fine-tuning details
Llama-231 fine-tunes were performed using a modified version of the Meta Research Llama-2 recipes repository; the modified repository can be found at https://github.com/lbnlp/nerre-llama. Llama-2 fine-tunes were performed using the 70 billion parameter version of Llama-2 (llama-2-70b-hf) with quantization (8 bit precision). The number of epochs was set to 7 for doping tasks and 4 for the MOF/general tasks. Llama-2 fine-tunes used parameter efficient fine-tuning (PEFT) using low rank adaptation (LoRA)58 with LoRA r = 8, α = 32 and LoRA dropout of 0.05. Further hyperparameter tuning was not performed. Decoding was done without sampling using greedy decoding to be consistent with GPT-3 decoding setting of temperature = 0, with max tokens = 512 for doping task and 1024 for general and MOF task. More details on the fine tuning and inference parameters are available in the modified repository and Supplementary Note 3. All fine-tuning and inference was performed on a single A100 (Ampere) tensor core GPU with 80GB VRAM.
The fine-tuned weights for each model are provided in the NERRE-Llama repository (url above) along with code and instructions for downloading the weights, instantiating the models, and running inference.
Evaluation criteria
The fuzzy and complex nature of the entities and relationships detailed in the previous section necessitates the use of several metrics for scoring. We evaluate the performance of all models on two levels:
-
1.
A relation F1 computed on a stringent exact word-match basis (i.e., how many words are correctly linked together exactly as they appear in the source text prompt).
-
2.
A holistic information extraction F1 based on manual inspection by a domain expert, which doesn’t require words to match exactly.
We separately provide a sequence-level error analysis in Supplementary Note 7 and Supplementary Discussion 1.
NERRE performance
We measure NERRE performance as the ability of the model to jointly recognize entities and the relationships between them.
Exact word-match basis scoring
We score named entity relationships on a word-basis by first converting an entity E into a set of constituent k whitespace-separated words E = {w1, w2, w3, …, wk}. When comparing two entities Etrue and Etest that do not contain chemical formulae, we count the number of exactly matching words in both sets as true positives (Etrue ∩ Etest) and the mathematical set differences between the sets as false positives (Etest − Etrue) or false negatives (Etrue − Etest). For example, if the true entity is “Bi2Te3 thin film” and the predicted entity is “Bi2Te3 film sample”, we record two true positive word exact matches (“Bi2Te3″, “film”), one false negative (“thin”), and one false positive (“sample”). Formula-type entities are crucial for identifying materials, so in cases where entities contain chemical formulae, Etest must contain all wi that can be recognized as stoichiometries for any of wi ∈ Etest to be considered correct. For example, if the true entity is “Bi2Te3 thin film”, and the predicted entity is “thin film”, we record three false negatives. Thus, any formula-type entity (Doping host, Doping dopant, General formula, and MOF mof_formula) containing a chemical composition is entirely incorrect if the composition is not an exact match. This choice of evaluation was made to avoid metrics measuring the performance of the model in a misleading way. For example, “Bi2Te3 nanoparticles” and “Bi2Se3 nanoparticles” have very high similarities via Jaro-Winkler (0.977) and character-level BLEU-4 (0.858), but these two phrases mean entirely different things—the material’s chemistry is wrong. Under our scoring system, they are recorded as entirely incorrect because the compositions do not match.
We score relationships between entities on a word-by-word basis to determine the number of correct relation triplets. Triplets are 3-tuples relating word \({w}_{j}^{n}\) of an entity En to word \({w}_{k}^{m}\) of an entity Em by relationship r, represented as \(({w}_{j}^{n},{w}_{k}^{m},r)\). The total set of correct relationships Ttrue for a text contains many of these triplets. A test set of relationships Ttest is evaluated by computing the number of triplets found in both sets (Ttrue ∩ Ttest) as true positives and the differences between these sets as false positives (Ttest − Ttrue) or false negatives (Ttrue − Ttest). Entity triplets are also bound to the same requirement for composition correctness if either of the words in the triplet belong to an formula-type entity (host, dopant, formula, mof_formula), i.e., we count all triplets for two entities as incorrect if the formula is not an exact string match. With correct and incorrect triplets identified, F1 scores for each relation are calculated as:
$$\,{{\mbox{precision}}}=\frac{{{\mbox{No. of correct relations retrieved}}}}{{{\mbox{No. of relations retrieved}}}\,}$$
(1)
$$\,{{\mbox{recall}}}=\frac{{{\mbox{No. of correct relations retrieved}}}}{{{\mbox{No. of relations in test set}}}\,}$$
(2)
$${F}_{1}=\frac{2(\,{{\mbox{precision}}}\cdot {{\mbox{recall}}})}{{{\mbox{precision}}}+{{\mbox{recall}}}\,}$$
(3)
To compute triplet scores across entire test sets in practice, we first select a subset of relations to evaluate. We note that this is not a full evaluation of the task we are training the model to perform, which involves linking many interrelated entities simultaneously, but is rather provided to help give a general sense of its performance compared to other NERRE methods. For the doping task, we evaluate host–dopant relationships. For the general materials and MOF tasks, we evaluate relationships between the formula field (formula for general materials, mof_formula for MOFs) and all other remaining fields. For description, structure_or_phase, and applications fields, all of which may contain multiple values, all of the possible formula-value pairs are evaluated.
Manual evaluation
The metrics provided in prior sections demonstrate automatic and relatively strict methods for scoring NERRE tasks, but the underlying capabilities of the LLM models are best shown with manual evaluation. This is most apparent in the case of the General-JSON model, where exact boundaries on entities are fuzzier, precise definitions are difficult to universally define, and annotations include some implicit entity normalization. For example, the text “Pd ions were intercalated into mesoporous silica” may have equivalently have a correct description field for the material “silica” including “Pd-intercalated”, “Pd ion-intercalated”, “intercalated with Pd ions”, etc.; the exact choice of which particular string is used as the “correct” answer is arbitrary.
To better address scoring of these fuzzy tasks, we introduce an adjusted score based on a domain expert’s manual evaluation of whether the information extracted is a valid representation of the information actually contained in the passage. We term this adjusted score “manual score”; it constitutes a basis for precision, recall, and F1 that quantifies the quality of overall information capture for cases where there may be equivalent or multiple ways of representing the same concept. This score was constructed to better estimate the performance of our model for practical materials information extraction tasks.
We score entities extracted by annotators but not present in the model’s output as false negatives, except when reasonable variations are present. The criteria for a true positive are as follows:
-
1.
The entity comes from the original passage or is a reasonable variation of the entity in the passage (e.g., “silicon” ⟶ “Si”). It is not invented by the model.
-
2.
The entity is a root entity or is grouped with a valid root entity. For the General-JSON model, a root entity is either a material’s formula or name. If both are present, the formula is used at the root.
-
3.
The entity is in the correct field in the correct root entity’s group (JSON object).
Manual scores are reported per-entity as if they were NER scores. However, the requirements for a true positive implicitly include relational information, since an entity is only correct if is grouped with the correct root entity.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.